Center for Proteomics and Metabolomics
Glycomics
Proteomics
Metabolomics
MS Imaging
Bioinformatica
Promotietraject
Wat doet het CPM
Binnen het CPM leiden wij studenten met een Master diploma verder op tot volwaardige onderzoekers middels een promotietraject.
Een promotietraject binnen het LUMC betreft het volbrengen van een volledig onderzoeksproject (in een periode van 4 jaar), dat uitmondt in een proefschrift en het behalen van een doctorsgraad.
Bij het CPM verrichten onze promovendi onderzoek met behulp van de nieuwste technologieën, vaak in samenwerking met andere afdelingen van het LUMC, om een bijdrage te leveren aan het beter begrijpen en bestrijden van ziektes. Wij trainen onze promovendi o.a. in het correct uitvoeren van onderzoek, rapporteren, presenteren, een leven lang leren en innoveren.
…Wat doet het CPM
Binnen het CPM leiden wij studenten met een Master diploma verder op tot volwaardige onderzoekers middels een promotietraject.
Een promotietraject binnen het LUMC betreft het volbrengen van een volledig onderzoeksproject (in een periode van 4 jaar), dat uitmondt in een proefschrift en het behalen van een doctorsgraad.
Bij het CPM verrichten onze promovendi onderzoek met behulp van de nieuwste technologieën, vaak in samenwerking met andere afdelingen van het LUMC, om een bijdrage te leveren aan het beter begrijpen en bestrijden van ziektes. Wij trainen onze promovendi o.a. in het correct uitvoeren van onderzoek, rapporteren, presenteren, een leven lang leren en innoveren.
Promovendi worden gefinancierd door fondsen of particuliere giften. De promovendus werkt onder begeleiding van een promotor (hoogleraar) en eventuele medebegeleiders, de zogenaamde co-promotor(en).
Wil je meer informatie over promoveren binnen het LUMC?
Bezoek dan hier de website van de Gradute school.
Alle openstaande vacatures kun je vinden op de Werkenbij pagina of op de vacaturepagina van het LUMC.
Werken bij
Zie hier onze actuele vacatures.
Stagiaires en gastonderzoek
Naast het reguliere onderwijs verzorgt het CPM ook onderwijs in de vorm van stages en begeleiding/opleiding van gastonderzoekers.
Stages
Bij het CPM zijn er regelmatig mogelijkheden voor studenten om stage te lopen. Studenten met een bèta-opleiding die een carrière ambiëren in de biomedische wetenschappen, analytische chemie of bio-informatica, nodigen wij van harte uit om contact met ons op te nemen. In de regel gaan wij uit van een stageperiode van minstens 5 maanden.
Een lijst met reeds uitgevoerde stageprojecten bij de afdeling CPM is hier te vinden.
Naast het reguliere onderwijs verzorgt het CPM ook onderwijs in de vorm van stages en begeleiding/opleiding van gastonderzoekers.
Stages
Bij het CPM zijn er regelmatig mogelijkheden voor studenten om stage te lopen. Studenten met een bèta-opleiding die een carrière ambiëren in de biomedische wetenschappen, analytische chemie of bio-informatica, nodigen wij van harte uit om contact met ons op te nemen. In de regel gaan wij uit van een stageperiode van minstens 5 maanden.
Een lijst met reeds uitgevoerde stageprojecten bij de afdeling CPM is hier te vinden.
Gastonderzoekers
Het CPM begeleidt vaak onderzoekers van andere onderzoeksinstituten. Het doel van onze gastonderzoekers is o.a. het leren van technieken, of het uitvoeren van analyses op onze apparatuur, alsmede het testen van de reproduceerbaarheid van onderzoeksresultaten. Gastonderzoekers nodigen wij van harte uit om contact met ons op te nemen over de mogelijkheden.
Contactgegevens voor stages en gastonderzoek
Voor meer informatie en beschikbaarheid van plaatsing van stages verzoeken we je contact op te nemen met Dr. Paul Hensbergen. Stuur je CV en een motivatiebrief waarin je aangeeft wat je met je stage wilt bereiken naar: p.j.hensbergen@lumc.nl
Heb je interesse in een samenwerking met onze afdeling, neem dan contact op met ons afdelingshoofd Prof. dr. Manfred Wuhrer via m.wuhrer@lumc.nl en vermeld in je e-mail duidelijk wat je onderzoeksdoelen en je verwachtingen zijn.
Wat doet onze afdeling?
Proteomics en metabolomics: wat is dat?
In het menselijk lichaam zijn heel veel processen gaande. De meeste processen worden uitgevoerd door eiwitten. Eiwitkunde (proteomics) is de studie naar eiwitten in een organisme of deel van een organisme.
De stofwisseling in het menselijk lichaam is belangrijk om alle processen met energie te voorzien maar ook om bouwstenen aan te dragen. De studie naar stofwisselproducten of metabolieten wordt metabolomics genoemd. Een belangrijk onderwerp binnen de metabolomics is het zoeken naar patronen en afwijkingen in het metabolieten profiel die aanwijzingen geven over de gezondheidstoestand (ook wel biomarkers genoemd).
Proteomics en metabolomics: wat is dat?
In het menselijk lichaam zijn heel veel processen gaande. De meeste processen worden uitgevoerd door eiwitten. Eiwitkunde (proteomics) is de studie naar eiwitten in een organisme of deel van een organisme.
De stofwisseling in het menselijk lichaam is belangrijk om alle processen met energie te voorzien maar ook om bouwstenen aan te dragen. De studie naar stofwisselproducten of metabolieten wordt metabolomics genoemd. Een belangrijk onderwerp binnen de metabolomics is het zoeken naar patronen en afwijkingen in het metabolieten profiel die aanwijzingen geven over de gezondheidstoestand (ook wel biomarkers genoemd).
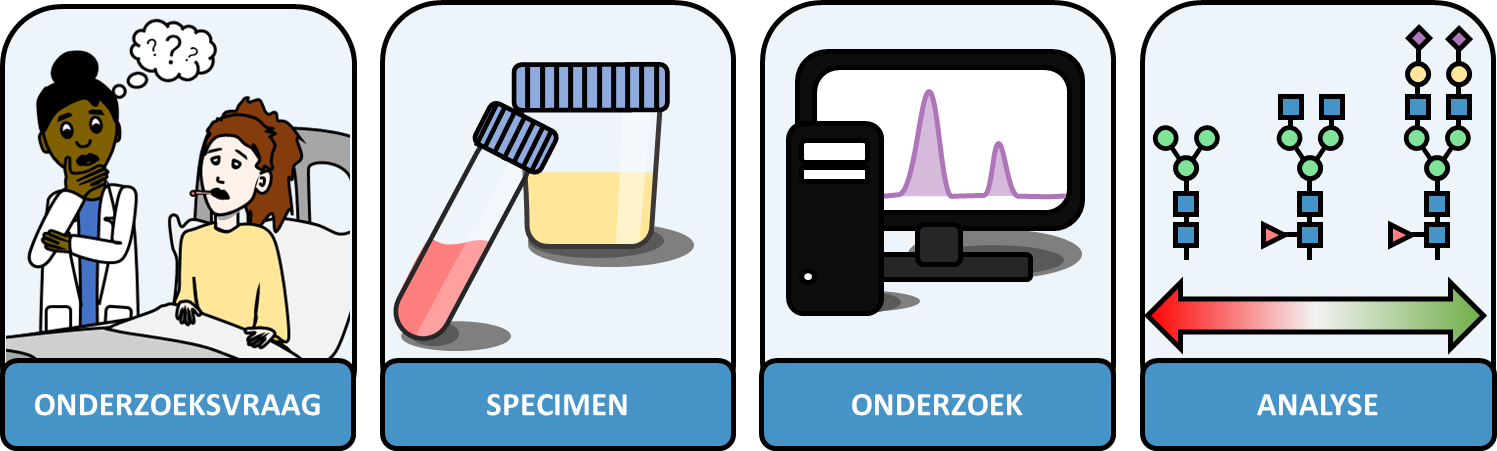
Het Centrum voor Proteomics en Metabolomics: wat doen wij?
Het Centrum voor Proteomics en Metabolomics (CPM) doet onderzoek naar eiwitten en metabolieten om ziekte-gerelateerde veranderingen in kaart te brengen. Dit onderzoek doen wij met behulp van o.a. massaspectrometers. Massaspectrometers zijn apparaten waarmee onderzoekers eiwitten of fragmenten ervan kunnen karakteriseren om daarmee biologische vragen te beantwoorden. Het CPM is opgedeeld in verschillende expertisegebieden:
Proteomics
De proteomics groep heeft als voornaamste doel methodes te ontwikkelen ter verbetering van de identificatie en charcterisatie van eiwitten, zodat een beter inzicht verkregen kan worden in ziekteprocessen.
Metabolomics
Binnen de metabolomics groep wordt onderzoek gedaan naar de ontwikkeling en toepassing van analysetechnieken voor onderzoek naar kleine moleculen zoals metabolieten en lipiden.
Glycomics
Glycanen (suikerketens) die aan een eiwit vastzitten kunnen veranderen van structuur en positie. Binnen de glycomics groep worden technieken ontwikkeld en toegepast om dergelijke veranderingen bij ziekteprocessen te bestuderen.
Mass Spectometry Imaging
Binnen de beeldvormende massaspectrometrie groep (Mass Spectometry Imaging Group) wordt microscopische beeldvorming gecombineerd met de analytische eigenschappen van een massaspectrometer. Een doel van deze groep is het ontwikkelen van nieuwe methoden om meer informatie over eiwitten, metabolieten en lipiden uit weefsels te halen. Een ander doel is om methodes toe te passen in pre-klinische studies, om pathologen, moleculair- en celbiologen van moleculaire informatie uit weefsel te voorzien ter ondersteuning van onderzoek naar ziekten processen.
Bioinformatics
Massa spectrometrische experimenten genereren een enorme berg aan data. De bioinformatica groep gebruikt computertechnieken om de data vanuit verschillende onderzoeken samen te brengen om zo humane ziektes beter te begrijpen.
Nieuws
Hier vindt u het laatste nieuws van het Center for Proteomics and Metabolomics.
February 2020
Osteoarchaeologist dr. Rachel Schats investigates traces of Malaria infection on human archaeologic skeletons under an NWO VENI grant.
At the Center for Proteomics and Metabolomics we assist her with the extraction and detection of hemozoin, a detergent of the malaria parasite, using mass spectrometry. After developing a robust method, samples of human remaining's from Middle Age Netherlands will be analysed, to create a geographic overview of Malaria infection during the Middle Age, a period in our history of which less is known concerning this topic.
…Hier vindt u het laatste nieuws van het Center for Proteomics and Metabolomics.
February 2020
Osteoarchaeologist dr. Rachel Schats investigates traces of Malaria infection on human archaeologic skeletons under an NWO VENI grant.
At the Center for Proteomics and Metabolomics we assist her with the extraction and detection of hemozoin, a detergent of the malaria parasite, using mass spectrometry. After developing a robust method, samples of human remaining's from Middle Age Netherlands will be analysed, to create a geographic overview of Malaria infection during the Middle Age, a period in our history of which less is known concerning this topic.
January 23, 2020
NWO LIFT project awarded: Proteoform-resolved pharmacokinetics of biopharmaceuticals
Lead applicant: Professor Manfred Wuhrer, LUMC
Consortium: LUMC, Roche Diagnostics GmbH
Modern medicines are often proteins administered into the bloodstream. These proteins will often undergo changes after this administration; for instance, they may lose part of their molecular structure, or this may become modified. The LUMC, working together with two international companies (Roche and Sciex), will devise methods to map the changes undergone by administered proteins, and then examine the influence these changes have, not just on the lifetime of the drug but also on its activity. The project will contribute towards quality control, improved drug applicability, and the development of new medicines.
https://www.nwo.nl/en/news-and-events/news/2020/01/green-light-for-eleven-public-private-lift-research-projects-html
October 3, 2019
NWO KLEIN-1 project awarded: Cleaning like a Pro:PPEP (Pro-Pro endopeptidase)-regulated processes in Clostridiodes difficile
Dr. P.J. Hensbergen, LUMC
Clostridioides difficile , a bacterium that causes intestinal infections, secretes an enzyme (PPEP-1) that regulates the spread of the bacterium. In doing so it cleaves surface proteins from the bacteria, proteins which are necessary for attachment. However, in the case of one of these surface proteins, we have yet to discover what it attaches to. PPEPs also occur in other bacteria, but their activity is slightly different. C. difficile also has a second PPEP, but its function is unknown. In this project we want to further unpack the PPEP-regulated processes in C. difficile, and to better understand the activity of different PPEPs.
Publicaties
Ons Team
- Prof. dr. Manfred Wuhrer (Head of the Center for Proteomics and Metabolomics)
- Proteomics and Glycomics
- Dr. Peter van Veelen (Head Proteomics group)
- Proteomics
- Dr. Martin Giera (Head Metabolomics group)
- Metabolomics
- Prof. dr. Manfred Wuhrer en Dr. Martin Giera
- Mass Spectrometry Imaging
- Dr. Magnus Palmblad (Head Bioinformatics)
- Mass Spectrometry
- Proteomics
- Bioinformatics
Contactgegevens
Onze labs en kantoren zijn gevestigd in gebouw 1 van het Leids Universitair Medisch Centrum.
Bezoekadres:
LUMC
Afdeling Center for Proteomics and Metabolomics (CPM)
Kantoor P-01-064, gebouw 1 (route 920)
2333 ZA Leiden
Postadres:
LUMC
Afdeling Center for Proteomics and Metabolomics (CPM)
Postadres P1-Q
Postbus 9600
2300 RC Leiden
Voor meer informatie of vragen kunt u contact opnemen met:
Prof. dr. Manfred Wuhrer, Afdelingshoofd Center for Proteomics and Metabolomics
Tel: +31 71 526 6989
m.wuhrer@lumc.nl
Onze labs en kantoren zijn gevestigd in gebouw 1 van het Leids Universitair Medisch Centrum.
Bezoekadres:
LUMC
Afdeling Center for Proteomics and Metabolomics (CPM)
Kantoor P-01-064, gebouw 1 (route 920)
2333 ZA Leiden
Postadres:
LUMC
Afdeling Center for Proteomics and Metabolomics (CPM)
Postadres P1-Q
Postbus 9600
2300 RC Leiden
Voor meer informatie of vragen kunt u contact opnemen met:
Prof. dr. Manfred Wuhrer, Afdelingshoofd Center for Proteomics and Metabolomics
Tel: +31 71 526 6989
m.wuhrer@lumc.nl
Management assistent: Riemke Louwerse - Siemons
Tel: +31 71 526 6982
cpmsecretary@lumc.nl
Projectmanager: Suzanne van der Plas- Duivesteijn
Tel: +31 71 526 6984
S.J.Duivesteijn@lumc.n