Characterisation of pluripotency status, functional pluripotency and cell identity
Costs per clone:
LUMC External academia
…Costs per clone:
LUMC External academia
Pluripotency status, morphology,
functional pluripotency, STR €82 €192
Karyotyping
ddPCR assay StemGenomics* €209 €345
G-banding** €253 €376
* ICS digital PSC 24 probes assay; www. stemgenomics.com
** 20 metaphases
Characterisation of hiPSCs for quality control (QC)
hiPSC lines are characterised as follows:
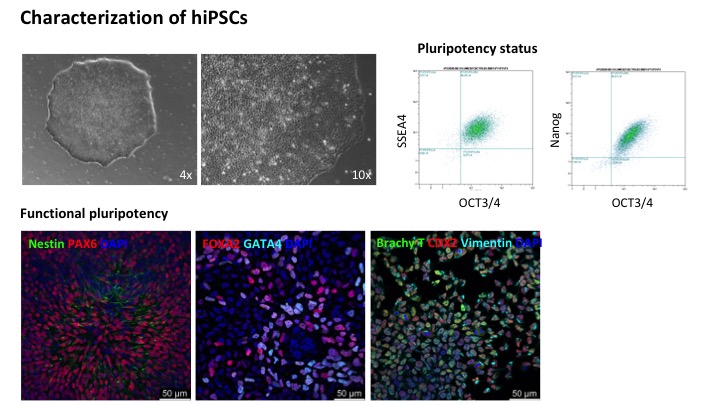
Morphology
Brightfield images of representative hiPSC colonies.
Pluripotency status
The percentage of undifferentiated cells expressing SSEA4, Nanog and OCT3/4 will be determined by FACS analysis. hiPSCs pass QC when ≥ 70% express SSEA4/Nanog and SSEA4/OCT3/4.
Functional pluripotency
hiPSCs are differentiated short-term using the STEMdiff Trilineage Differentiation Kit (STEMCELL Technologies). The presence of derivatives of ecto-, endo- and mesoderm is analysed by immunofluorescent (IF) staining with 3 markers per germ layer: Pax6, Nestin, FABP7 (ectoderm); CDX2, Vimentin, Brachyury T (mesoderm); FOXA2, GATA4, EOMES (endoderm). hiPSCs pass QC when at least 2 markers per germ layer are present.
…hiPSC lines are characterised as follows:
Morphology
Brightfield images of representative hiPSC colonies.
Pluripotency status
The percentage of undifferentiated cells expressing SSEA4, Nanog and OCT3/4 will be determined by FACS analysis. hiPSCs pass QC when ≥ 70% express SSEA4/Nanog and SSEA4/OCT3/4.
Functional pluripotency
hiPSCs are differentiated short-term using the STEMdiff Trilineage Differentiation Kit (STEMCELL Technologies). The presence of derivatives of ecto-, endo- and mesoderm is analysed by immunofluorescent (IF) staining with 3 markers per germ layer: Pax6, Nestin, FABP7 (ectoderm); CDX2, Vimentin, Brachyury T (mesoderm); FOXA2, GATA4, EOMES (endoderm). hiPSCs pass QC when at least 2 markers per germ layer are present.
Cell line authentication
One hiPSC line per donor sample and the corresponding primary cells are subjected to short tandem repeat (STR) analysis to confirm matching identity. This service is performed by the LUMC department of Human Genetics.
Upon completion of characterisation the customer is provided with:
- Cryovials for each characterised hiPSC line at 2 passages (3 vials each)
- Cryovials for each uncharacterised hiPSC line at 1 passage (2 vials)
- Summary of characterisation/ QC data (PowerPoint; raw data upon request)
Standard Operation Procedure (SOP) for starting up cells
&w=826&h=464)