Genome instability and genome engineering
The information is organized in 23 pairs of chromosomes and written in a language that only has 4 letters: the bases G, A, T and C. Each human cell contains 12 billion of these letters. Together, the 46 chromosomes are referred to as the human genome - the genome, defined as the complement of genetic material that makes up an organism. Genome stability is essential to accurately transmit all genetic information to progeny (or daughter cells upon cell division) to safeguard genetic integrity and prevent disease. Unfortunately, nevertheless, all of us will (personally, or in the people surrounding us) experience the fact that this process is not without error. Mutations resulting from errors during DNA replication, or resulting from copying or mis-repair of damaged DNA will accumulate during our lifetime and cause the disease that is responsible for a quarter of all deaths in the Western world: cancer.
…The information is organized in 23 pairs of chromosomes and written in a language that only has 4 letters: the bases G, A, T and C. Each human cell contains 12 billion of these letters. Together, the 46 chromosomes are referred to as the human genome - the genome, defined as the complement of genetic material that makes up an organism. Genome stability is essential to accurately transmit all genetic information to progeny (or daughter cells upon cell division) to safeguard genetic integrity and prevent disease. Unfortunately, nevertheless, all of us will (personally, or in the people surrounding us) experience the fact that this process is not without error. Mutations resulting from errors during DNA replication, or resulting from copying or mis-repair of damaged DNA will accumulate during our lifetime and cause the disease that is responsible for a quarter of all deaths in the Western world: cancer.
Our research focusses on understanding and exploiting the mechanisms that prevent or cause genome instability and mutagenesis. We particularly study how cells deal with obstructions to efficient and accurate DNA replication, such as thermodynamically stable secondary structures (e.g. G-quadruplexes) and damaged bases. These obstacles can lead to chromosomal breaks, which when left unrepaired cause cell death and/or aneuploidy, but in case of inaccurate repair drive carcinogenesis.
Key discoveries we recently made are:
- Cells employ an alternative mechanism to repair DNA breaks that result from replication fork obstacles, which we termed polymerase Theta-Mediated End Joining (TMEJ), as it critically depends on the functionality of the A-family polymerase Theta(Koole et al., Nature Commun. 2014; Roerink et al., Genome Res. 2014; Lemmens et al., Nature Commun. 2105; van Schendel et al., PLOS Genet. 2016).
- TMEJ is a key driver of random integration of DNA in mammalian cells, a phenomenon also known as illegitimate recombination(Zelensky et al., Nature Commun. 2017).
- We identified the mechanism by which plant cells incorporate foreign DNA into their genome, a process underlying transgenesis (Van Kregten et al., Nature Plants 2016). At the Institute Biology Leiden (IBL), we now further exploit the knowledge of DNA repair mechanisms in genome engineering strategies, with a special focus on plant biotechnology and crop development.
- Different repair pathways act to repair CRISPR-induced DNA breaks (van Schendel et al., Nature Commun. 2015; Schimmel et al., EMBO J. 2017)
Key discoveries we recently made are:
- Cells employ an alternative mechanism to repair DNA breaks that result from replication fork obstacles, which we termed polymerase Theta-Mediated End Joining (TMEJ), as it critically depends on the functionality of the A-family polymerase Theta(Koole et al., Nature Commun. 2014; Roerink et al., Genome Res. 2014; Lemmens et al., Nature Commun. 2105; van Schendel et al., PLOS Genet. 2016).
- TMEJ is a key driver of random integration of DNA in mammalian cells, a phenomenon also known as illegitimate recombination(Zelensky et al., Nature Commun. 2017).
- We identified the mechanism by which plant cells incorporate foreign DNA into their genome, a process underlying transgenesis (Van Kregten et al., Nature Plants 2016). At the Institute Biology Leiden (IBL), we now further exploit the knowledge of DNA repair mechanisms in genome engineering strategies, with a special focus on plant biotechnology and crop development.
- Different repair pathways act to repair CRISPR-induced DNA breaks (van Schendel et al., Nature Commun. 2015; Schimmel et al., EMBO J. 2017)
While the primary aim of this basic research is to provide mechanistic insight into the processes that are responsible for the maintenance of genome integrity and how these processes protect against tumor development, a number of clinically relevant implications have been identified for cancer as well as for gene-correction applications. Also these are subject of ongoing investigation.
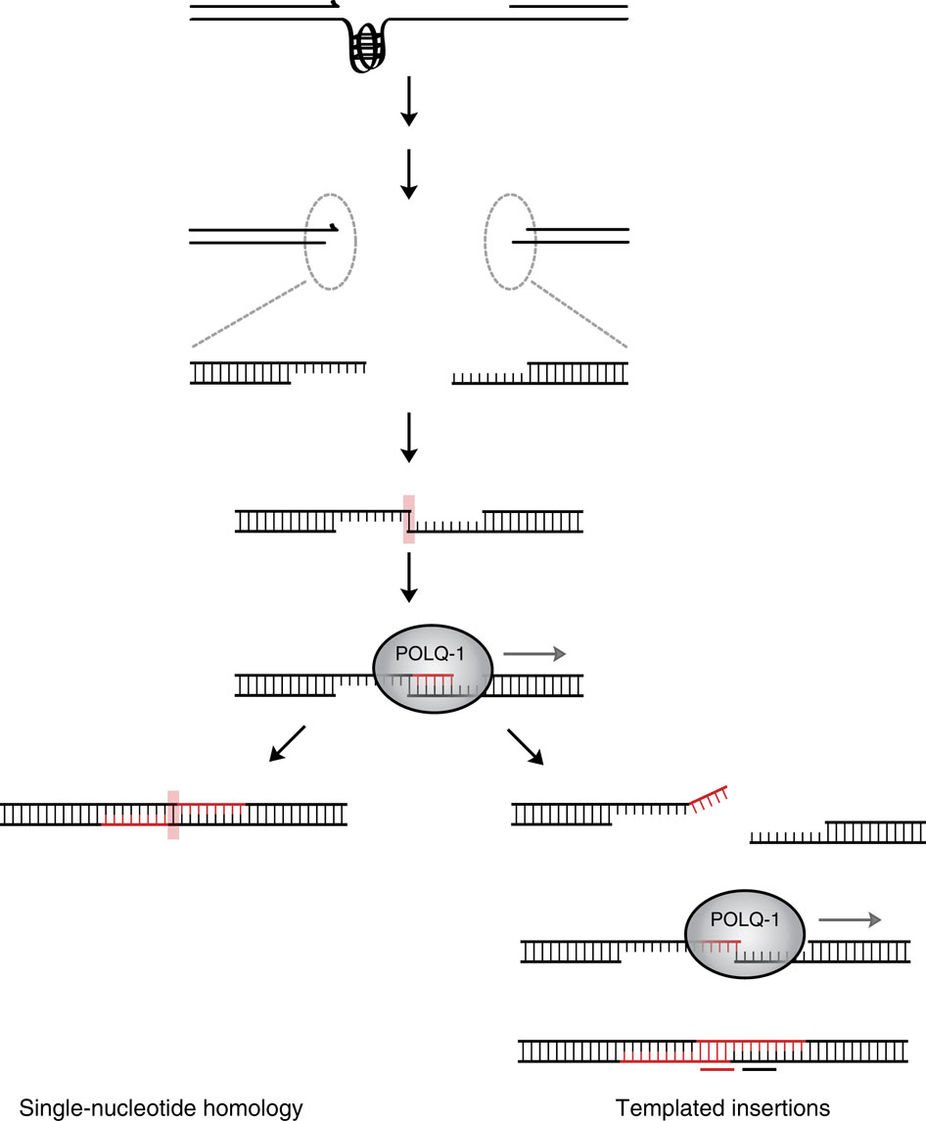
Figure 1. A replication fork block at a G4 structure results in a DSB. The broken ends are joined by polymerase Theta-mediated end joining (TMEJ) resulting in two types of deletions: first, simple ones characterized by single-nucleotide homology, and secondly, deletions with associated templated insertions. The generation of the latter class is simply an iteration of the steps leading to the simple deletions, with one exception being the dissociation of both ends after initial Pol Theta-mediated templated DNA synthesis.
Key publications
Our Team
Prof.dr. Marcel Tijsterman
Principal Investigator / Professor Genome Stability
Robin van Schendel
Senior researcher
Joost Schimmel
Senior researcher
Marco Barazas
Researcher
Prof.dr. Marcel Tijsterman
Principal Investigator / Professor Genome Stability
Robin van Schendel
Senior researcher
Joost Schimmel
Senior researcher
Marco Barazas
Researcher
Jan Boei
Researcher
Lejon E.M. Kralemann
Researcher (IBL)
Anna Dekker
PhD student
Marloes D. van Wezel
PhD student
Hanneke M. Kool
Research technician