Generation of research-grade human induced pluripotent stem cells (hiPSCs)
Note: All LUMC researchers must have a hiPSC research protocol approved by the LUMC ethical committee.
For commercial use, customers must obtain a license for the reprogramming vector.
A short project description (disease, mutation(s), donor sex, year of birth, goal of study; only for internal use by the LUMC hiPSC Hotel) is required for each request.
Please submit your request via the website Divvly www.divvly.com.
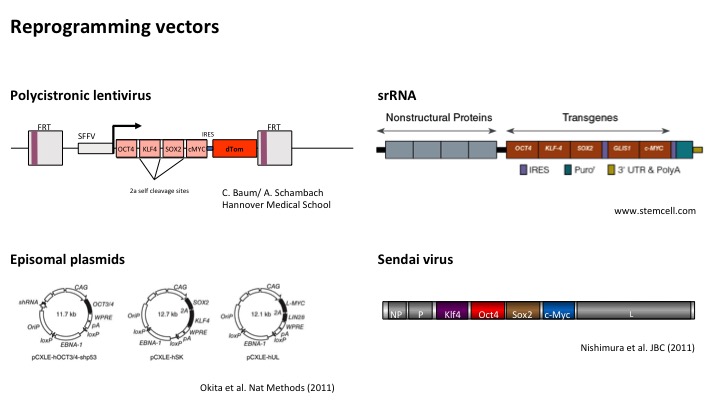
The LUMC hiPSC Hotel offers the generation of hiPSCs with the following vector systems:
Polycistronic lentivirus (LV)
For reprogramming of skin fibroblasts, erythroblasts, BOECs, renal epithelial and dental pulp cells. Integrates into the host genome. FLP sites flanking the transgenes can be used for excision by FLP enzyme (not included in the service).
Episomal plasmids
For reprogramming of skin fibroblasts, erythroblasts and renal epithelial cells. Mostly non-integrating but occasional (partial) integrations have been reported. Screening for integrations is not included in the service.
…The LUMC hiPSC Hotel offers the generation of hiPSCs with the following vector systems:
Polycistronic lentivirus (LV)
For reprogramming of skin fibroblasts, erythroblasts, BOECs, renal epithelial and dental pulp cells. Integrates into the host genome. FLP sites flanking the transgenes can be used for excision by FLP enzyme (not included in the service).
Episomal plasmids
For reprogramming of skin fibroblasts, erythroblasts and renal epithelial cells. Mostly non-integrating but occasional (partial) integrations have been reported. Screening for integrations is not included in the service.
Sendai virus (SeV)
For reprogramming skin fibroblasts and renal epithelial cells. Non-integrating. SeV-hiPS cells have to be maintained at the ML-2 safety level until the absence of SeV has been confirmed by qPCR and IF staining. The LUMC hiPSC Hotel checks for the absence of SeV in up to 3 clones at the passage indicated to the customer. Customers are responsible for testing for the absence of SeV used at a lower passage and/or in untested clones. Protocols for SeV detection are available upon request.
Self-replicating (sr)RNA
We use srRNA for reprogramming skin fibroblasts and renal epithelial cells. The resulting hiPSCs are integration-free.
In general, a minimum of 3 and a maximum of 6 hiPSC colonies (‘clones’) are picked for each donor sample; 2 reprogramming rounds are performed if necessary. Clones are expanded towards a hiPSC line and cryopreserved at 2 passages (usually P3-6, characaterized clones) and 1 passage (uncharacterized clones).
Note: We cannot guarantee that hiPSC lines are truly clonal, that is that they originated from a single cell.
Costs services
Costs of our services: Isolation of primary cells from patient tissues, temporary banking, mycoplasma testing and generation of research grade human induced pluripotent stem cells (hiPSCs) per sample (excl. shipping and VAT). Costs for characterization are indicated below.
Costs per donor sample:
| LUMC | External academia | |
|---|---|---|
| Fibroblasts/ SeV | upon request | upon request |
| Fibroblasts/ LV | €1360 | €3310 |
| Fibroblasts/ episomal | €1370 | €3320 |
| Fibroblasts/ srRNA | €1680 | €3650 |
| LUMC | External academia | |
|---|---|---|
| Fibroblasts/ SeV | upon request | upon request |
| Fibroblasts/ LV | €1360 | €3310 |
| Fibroblasts/ episomal | €1370 | €3320 |
| Fibroblasts/ srRNA | €1680 | €3650 |
| Erythroblasts/ episomal | €1710 | €3685 |
| Renal epithelium/ srRNA | €1840 | €3815 |
| Costs for other combinations and commercial customers upon request. |
&w=826&h=464)